using Pumas
using PumasUtilities
using Random
using AlgebraOfGraphics
using CairoMakie
using CSV
using DataFramesMeta
PK03 - One-compartment 1st and 0-order input
1 Objectives
In this tutorial, we will be looking at building a one compartment model for zero-order input and simulating the model for 1 subject.
2 Background
- Structural model - One compartment linear elimination with zero-order absorption
- Route of administration - Oral
- Dosage Regimen - 20 mg Oral
- Number of Subjects - 1
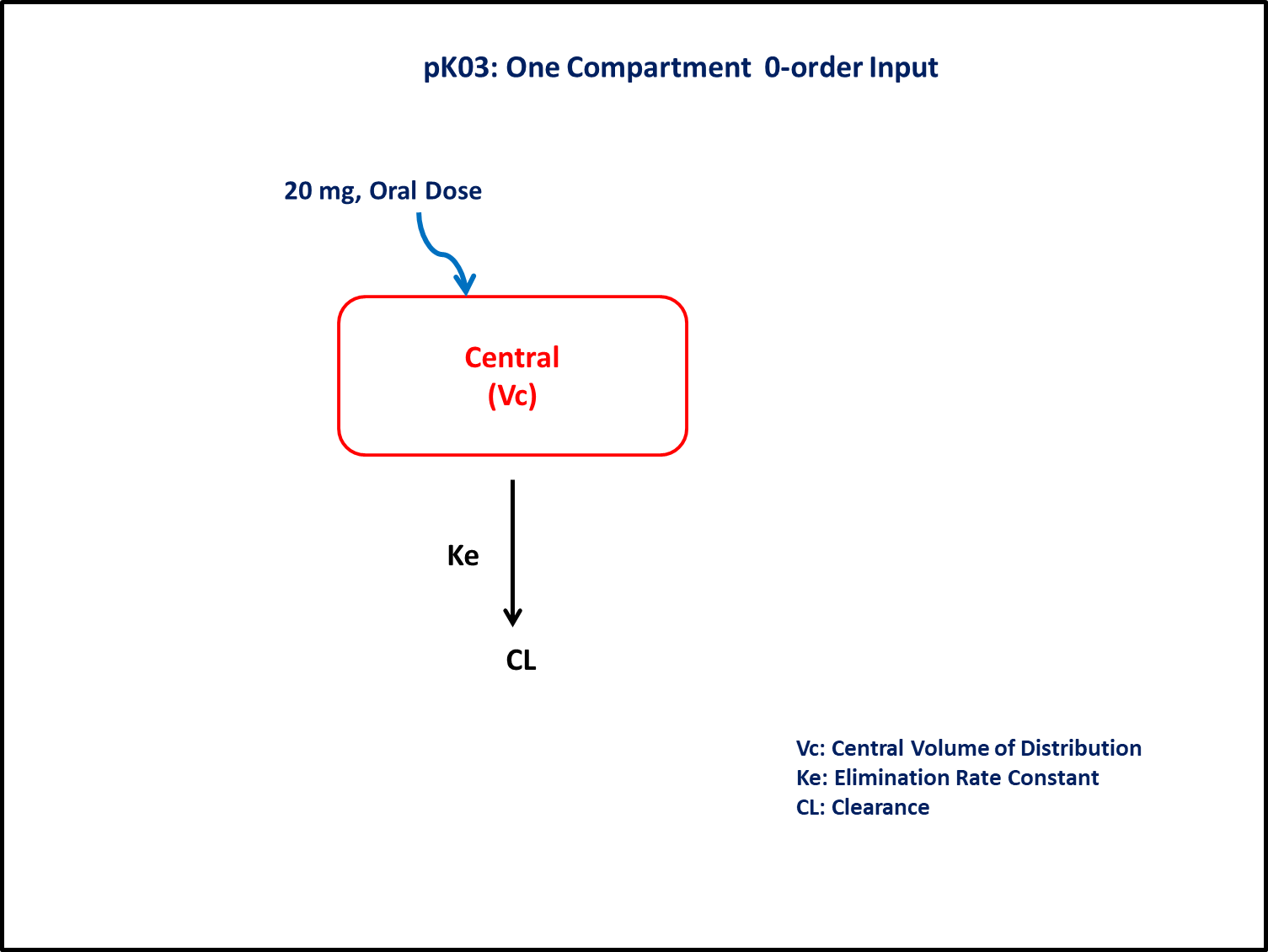
In this model, a collection of plasma concentration data will help us to derive/estimate the following parameters: Clearance, Volume of Distribution, and Duration of zero-order input.
3 Libraries
Call the required libraries to get started.
4 Model
In this one compartment model, we administer the dose in the Central compartment as a zero-order input and estimate the rate of input.
pk_03 = @model begin
@metadata begin
desc = "One Compartment Model with zero-order input"
timeu = u"hr"
end
@param begin
"""
Clearance (L/hr)
"""
tvcl ∈ RealDomain(lower = 0)
"""
Volume (L)
"""
tvvc ∈ RealDomain(lower = 0)
"""
Assumed Duration of Zero-order (hr)
"""
tvTabs ∈ RealDomain(lower = 0)
Ω ∈ PDiagDomain(3)
"""
Proportional RUV
"""
σ²_prop ∈ RealDomain(lower = 0)
end
@random begin
η ~ MvNormal(Ω)
end
@pre begin
Cl = tvcl * exp(η[1])
Vc = tvvc * exp(η[2])
end
@dosecontrol begin
duration = (Central = tvTabs * exp(η[3]),)
end
@dynamics begin
Central' = -(Cl / Vc) * Central
end
@derived begin
"""
PK03 Concentration (μg/L)
"""
cp = @. 1000 * (Central / Vc)
"""
PK03 Concentration (μg/L)
"""
dv ~ @. Normal(cp, sqrt(cp^2 * σ²_prop))
end
endPumasModel
Parameters: tvcl, tvvc, tvTabs, Ω, σ²_prop
Random effects: η
Covariates:
Dynamical system variables: Central
Dynamical system type: Matrix exponential
Derived: cp, dv
Observed: cp, dv5 Parameters
Parameters provided for simulation:
Cl- Clearance (L/hr)Vc- Volume of Central Compartment (L)Tabs- Assumed duration of zero-order input (hrs)Ω- Between Subject Variabilityσ- Residual error
These are the initial estimates we will be using in this model exercise. tv represents the typical value for parameters.
param_03 = (;
tvcl = 45.12,
tvvc = 96,
tvTabs = 4.54,
Ω = Diagonal([0.08, 0.03, 0.0226]),
σ²_prop = 0.015,
)6 Dosage Regimen
Single 20 mg or 20000 μg oral dose given to a subject.
In this the dose administered is on mg and conc are in μg/L, hence a scaling factor of 1000 is used in the @derived block in the model.
ev1 = DosageRegimen(20; rate = -2)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 1 | 20.0 | 1 | 0.0 | 0 | -2.0 | 0.0 | 0 | NullRoute |
sub = Subject(; id = 1, events = ev1)Subject
ID: 1
Events: 17 Simulation
Let’s simulate for plasma concentration with the specific observation time points after oral administration.
The Random.seed! function is included here for purposes of reproducibility of the simulation in this tutorial. Specification of a seed value would not be required in a Pumas workflow that is estimating model parameters.
Random.seed!(123)The random effects are zero’ed out since we are simulating means
zfx = zero_randeffs(pk_03, sub, param_03)sim = simobs(pk_03, sub, param_03, zfx, obstimes = 0:0.1:10)SimulatedObservations
Simulated variables: cp, dv
Time: 0.0:0.1:10.08 Visualize results
@chain DataFrame(sim) begin
dropmissing(:cp)
data(_) *
mapping(:time => "Time (hours)", :cp => "PK03 Concentration (μg/L)") *
visual(Lines, linewidth = 4)
draw(; axis = (; xticks = 0:1:10,), figure = (; fontsize = 22))
end9 Population Simulation
This block updates the parameters of the model to increase intersubject variability in parameters and defines time points for the prediction of concentrations. The results are written to a CSV file.
par = (
tvcl = 45.12,
tvvc = 96,
tvTabs = 4.54,
Ω = Diagonal([0.09, 0.04, 0.0225]),
σ²_prop = 0.015,
)
ev1 = DosageRegimen(20; rate = -2)
pop = map(i -> Subject(id = i, events = ev1), 1:90)
Random.seed!(1234)
sim_pop = simobs(pk_03, pop, par, obstimes = [0, 0.5, 1, 1.5, 2, 3, 4, 5, 6, 7, 8, 9, 10])
df_sim = DataFrame(sim_pop);
#CSV.write("pk_03.csv", df_sim)