using PumasUtilities
using Random
using Pumas
using CairoMakie
using AlgebraOfGraphics
using CSV
using DataFramesMeta
using Dates
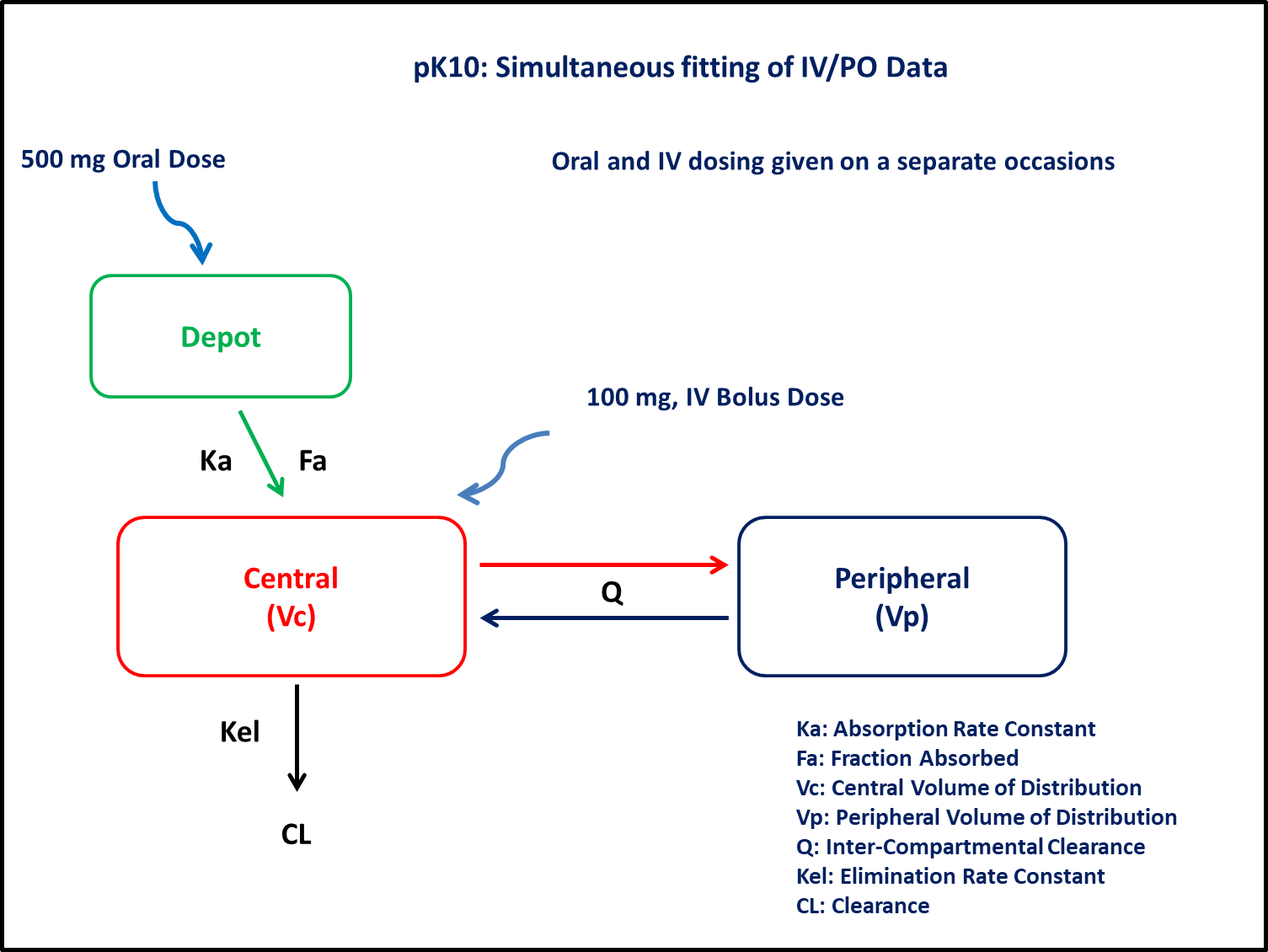
PK10 - Simultaneous fitting of IV/PO data
1 Background
- Structural model - Two compartment linear elimination with first order absorption
- Route of administration - IV bolus and oral given on separate occasions
- Dosage regimens - 100 mg IV Bolus and 500 mg Oral
- Subject - 1
2 Learning Outcome
This exercise demonstrates simultaneous fitting of IV/PO data and will help you understand the disposition of drugs following IV and oral administration (with and without lag time).
3 Objectives
To build a two-compartment model, simulate the model for a single subject given IV bolus and oral dose on separate occasions and subsequently perform simulation for a population.
4 Libraries
Load the necessary libraries.
5 Model definition
Note the expression of the model parameters with helpful comments. The model is expressed with differential equations. Residual variability is a proportional error model.
In this two compartment model, we administer doses to the Depot and Central compartments.
pk_10 = @model begin
@metadata begin
desc = "Two Compartment Model"
timeu = u"minute"
end
@param begin
"""
Volume of Central Compartment (L)
"""
tvvc ∈ RealDomain(lower = 0)
"""
Volume of Peripheral Compartment (L)
"""
tvvp ∈ RealDomain(lower = 0)
"""
InterCompartmental Clearance (L/min)
"""
tvq ∈ RealDomain(lower = 0)
"""
Clearance (L/min)
"""
tvcl ∈ RealDomain(lower = 0)
"""
Absorption Rate Constant (min⁻¹)
"""
tvka ∈ RealDomain(lower = 0)
"""
Fraction of drug absorbed
"""
tvfa ∈ RealDomain(lower = 0)
"""
Lagtime (min)
"""
tvlag ∈ RealDomain(lower = 0)
Ω ∈ PDiagDomain(7)
"""
Proportional RUV
"""
σ²_prop ∈ RealDomain(lower = 0)
end
@random begin
η ~ MvNormal(Ω)
end
@pre begin
Vc = tvvc * exp(η[1])
Vp = tvvp * exp(η[2])
Q = tvq * exp(η[3])
CL = tvcl * exp(η[4])
Ka = tvka * exp(η[5])
end
@dosecontrol begin
bioav = (Depot = tvfa * exp(η[6]),)
lags = (Depot = tvlag * exp(η[7]),)
end
@dynamics Depots1Central1Periph1
@derived begin
cp = @. Central / Vc
"""
Observed Concentration (mg/L)
"""
dv ~ @. Normal(cp, sqrt(cp^2 * σ²_prop))
end
endPumasModel
Parameters: tvvc, tvvp, tvq, tvcl, tvka, tvfa, tvlag, Ω, σ²_prop
Random effects: η
Covariates:
Dynamical system variables: Depot, Central, Peripheral
Dynamical system type: Closed form
Derived: cp, dv
Observed: cp, dv6 Initial Estimates of Model Parameters
The model parameters for simulation are the following. Note that tv represents the typical value for parameters.
Vc- Volume of Central Compartment (L)Vp- Volume of Peripheral Compartment (L)Q- InterCompartmental clearance (L/min)Cl- Clearance from Central InterCompartmental (L/min)Ka- Absorption rate constant (min⁻¹)Fa- Fraction of drug absorbedlags- Lagtime (min)Ω- Between Subject Variabilityσ- Residual error
6.1 IV / PO - without lagtime
A vector of model parameter values is defined.
param1 = (
tvvc = 59.9348,
tvvp = 60.5898,
tvq = 1.55421,
tvcl = 0.967573,
tvka = 0.0471557,
tvfa = 0.318748,
tvlag = 14.8187,
Ω = Diagonal([0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01]),
σ²_prop = 0.01,
)6.2 IV / PO - with lagtime
param2 = (param1..., tvlag = 14.8187)7 Dosage Regimen and Subjects
Dosage Regimen - single subject receiving 100 mg Intravenous bolus dose and 500 mg oral dose on different occasions.
7.1 IV
ev1 = DosageRegimen(100, time = 0, cmt = 2)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 2 | 100.0 | 1 | 0.0 | 0 | 0.0 | 0.0 | 0 | NullRoute |
sub1_iv = Subject(id = "ID:1 IV", events = ev1)Subject
ID: ID:1 IV
Events: 17.2 PO
ev2 = DosageRegimen(500, time = 0, cmt = 1)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 1 | 500.0 | 1 | 0.0 | 0 | 0.0 | 0.0 | 0 | NullRoute |
ids = ["ID:1 PO No Lag", "ID:1 PO With Lag"]
pop_po = map(i -> Subject(id = ids[i], events = ev2), 1:length(ids))Population
Subjects: 2
Observations: 8 Single-Subject Simulation
8.1 IV
Simulate plasma concentration with specific observation times after IV bolus.
Initialize the random number generator with a seed for reproducibility of the simulation.
Random.seed!(123)Define the timepoints at which concentration values will be simulated.
sim_iv_sub1 = simobs(pk_10, sub1_iv, param1, obstimes = 0.1:0.1:400)SimulatedObservations
Simulated variables: cp, dv
Time: 0.1:0.1:400.08.2 PO
Simulate plasma concentration with specific observation times after PO (with and without lag time)
sim_po_sub1 = map(zip(pop_po, [param1, param2])) do (subj, p)
return simobs(pk_10, subj, p, obstimes = 0.1:0.1:400)
endSimulated population (Vector{<:Subject})
Simulated subjects: 2
Simulated variables: cp, dv9 Visualize Results
all_sims = [sim_iv_sub1, sim_po_sub1...]
@chain DataFrame(all_sims) begin
dropmissing(:cp)
data(_) *
mapping(:time => "Time (minutes)", :cp => "Concentration (mg/L)"; color = :id => "") *
visual(Lines; linewidth = 4)
draw(;
axis = (; xticks = 0:50:400),
figure = (; fontsize = 22),
legend = (; position = :bottom),
)
end10 Perform a Population Simulation
We perform a population simulation with 50 participants.
This code demonstrates how to write the simulated concentrations to a comma separated file (.csv).
par = (
tvvc = 59.9348,
tvvp = 60.5898,
tvq = 1.55421,
tvcl = 0.967573,
tvka = 0.0471557,
tvfa = 0.318748,
tvlag = 14.8187,
Ω = Diagonal([0.04, 0.09, 0.0252, 0.0125, 0.06, 0.0225, 0.0158]),
σ²_prop = 0.0168738,
)
ev1 = DosageRegimen(100, time = 0, cmt = 2)
pop_iv = map(i -> Subject(id = i, events = ev1), 1:50)
Random.seed!(1234)
pop_sim_iv = simobs(pk_10, pop_iv, par, obstimes = 0:1:400)
df_pop_iv = DataFrame(pop_sim_iv)
df_pop_iv[!, :route] .= "IV"
ev2 = DosageRegimen(500, time = 0, cmt = 1)
pop_oral = map(i -> Subject(id = i, events = ev2), 1:50)
Random.seed!(1234)
pop_sim_oral = simobs(pk_10, pop_oral, par, obstimes = 0:1:400)
df_pop_oral = DataFrame(pop_sim_oral)
df_pop_oral[!, :route] .= "ORAL"
pkdata_10_sim = vcat(df_pop_iv, df_pop_oral)
#CSV.write("pk_10_sim.csv", pkdata_10_sim)11 Conclusion
This tutorial showed simultaneous fitting of IV/Oral data to understand the disposition of drugs.