using Random
using Pumas
using PumasUtilities
using AlgebraOfGraphics
using CairoMakie
using CSV
using DataFramesMeta
using Dates
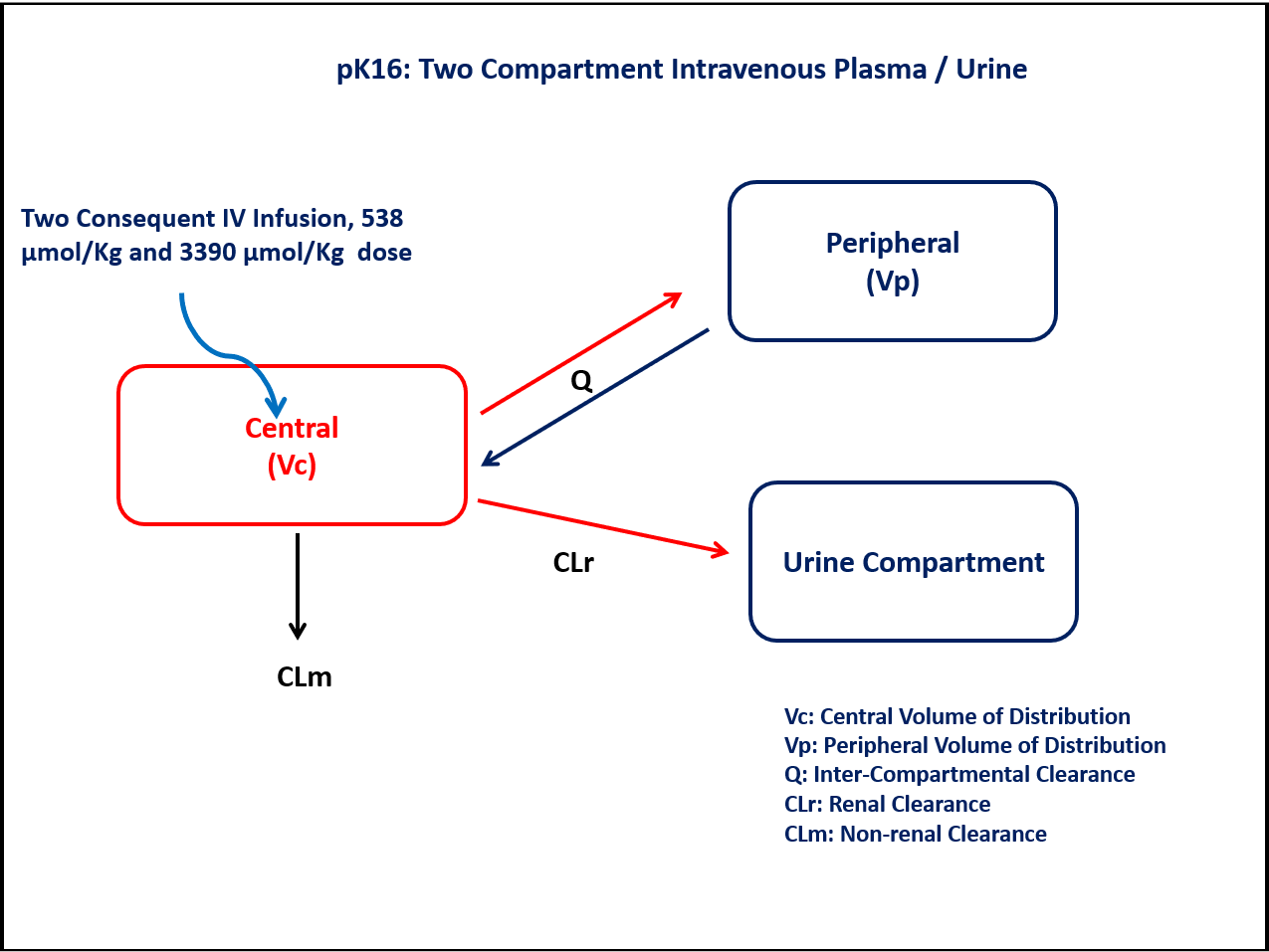
PK16 - Two-compartment intravenous plasma/urine
1 Background
- Structural Model - Two Compartment model with first order elimination
- Route of Administration - Intravenous infusion (Multiple-dose)
- Dosage Regimen - 538 µmol/kg followed by 3390 µmol/kg
- Number of Subjects - 1
2 Learning Outcome
This exercise explains a simultaneous analysis of plasma and urine data using a two-compartment model, with an additional urinary compartment accounting for the amount of drug excreted in urine.
3 Objectives
In this exercise, you will learn how to build a two compartment model and to simulate the data for a single subject based on the given dosage regimen.
4 Libraries
Call the necessary libraries to get started
5 Model
A two-compartment model with one Central and one Peripheral compartment will help in understanding the plasma concentration. A separate Urine compartment accounts for the fraction (amount) of drug excreted in urine.
pk_16 = @model begin
@metadata begin
desc = "Two Compartment Model with Urine Compartment"
timeu = u"hr"
end
@param begin
"""
Renal Clearance (L/hr/kg)
"""
tvclr ∈ RealDomain(lower = 0)
"""
Non-renal Clearance (L/hr/kg)
"""
tvclm ∈ RealDomain(lower = 0)
"""
Volume of Central Compartment (L/kg)
"""
tvvc ∈ RealDomain(lower = 0)
"""
Volume of Peripheral Compartment (L/kg)
"""
tvvp ∈ RealDomain(lower = 0)
"""
Inter-compartmental Clearance (L/hr/kg)
"""
tvq ∈ RealDomain(lower = 0)
Ω ∈ PDiagDomain(5)
"""
Proportional RUV - Plasma
"""
σ₁_prop ∈ RealDomain(lower = 0)
"""
Proportional RUV - Urine
"""
σ₂_prop ∈ RealDomain(lower = 0)
end
@random begin
η ~ MvNormal(Ω)
end
@pre begin
CLr = tvclr * exp(η[1])
CLm = tvclm * exp(η[2])
Vc = tvvc * exp(η[3])
Vp = tvvp * exp(η[4])
Q = tvq * exp(η[5])
CL = CLr + CLm
Vss = Vc + Vp
t12 = 0.693 * Vss / CL
end
@dynamics begin
Central' =
-(CLr / Vc) * Central - (CLm / Vc) * Central + (Q / Vp) * Peripheral -
(Q / Vc) * Central
Peripheral' = (Q / Vc) * Central - (Q / Vp) * Peripheral
Urine' = (CLr / Vc) * Central
end
@derived begin
cp_plasma = @. Central / (Vc + eps())
"""
Observed Plasma Concentration (μM)
"""
dv_plasma ~ @. Normal(cp_plasma, abs(cp_plasma) * σ₁_prop)
cp_urine = @. Urine
"""
Observed Urine Amount (μmol)
"""
dv_urine ~ @. Normal(cp_urine, abs(cp_urine) * σ₂_prop)
end
endPumasModel
Parameters: tvclr, tvclm, tvvc, tvvp, tvq, Ω, σ₁_prop, σ₂_prop
Random effects: η
Covariates:
Dynamical system variables: Central, Peripheral, Urine
Dynamical system type: Nonlinear ODE
Derived: cp_plasma, dv_plasma, cp_urine, dv_urine
Observed: cp_plasma, dv_plasma, cp_urine, dv_urine6 Parameters
CLr- Renal Clearance (L/hr/kg)CLm- Non-renal Clearance (L/hr/kg)Vc- Volume of Central Compartment (L/kg)Vp- Volume of Peripheral Compartment (L/kg)Q- Inter-compartmental Clearance (L/hr/kg)Ω- Between Subject Variabilityσ²₁- Residual error 1 (for plasma conc)σ²₂- Residual error 2 (for amount excreted in urine)
Derived Parameters:
CL- Total Clearance (L/hr/kg)Vss- Volume of distribution at steady state (L/kg)t1/2- Half life (hr)
Typical value (tv) estimates for individual parameters
param = (
tvclm = 0.05,
tvclr = 0.31,
tvvc = 1.6,
tvvp = 0.16,
tvq = 0.03,
Ω = Diagonal([0.04, 0.04, 0.04, 0.04, 0.04]),
σ₁_prop = 0.1,
σ₂_prop = 0.1,
)7 Dosage Regimen
- The subject (male dog) received two consecutive intravenous infusions of a drug.
- The subject received an initial dose of 538 µmol/kg from time 0 to 0.983 hr (rate = 547.304) followed by 3390 µmol/kg dose from time 0.983 to 23.95 hr (rate = 147.603).
- Total infused dose: 3928 µmol/kg.
ev1 = DosageRegimen([538, 3390], time = [0, 0.983], cmt = [1, 1], rate = [547.304, 147.603])| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 1 | 538.0 | 1 | 0.0 | 0 | 547.304 | 0.983 | 0 | NullRoute |
| 2 | 0.983 | 1 | 3390.0 | 1 | 0.0 | 0 | 147.603 | 22.967 | 0 | NullRoute |
sub1 = Subject(id = 1, events = ev1)Subject
ID: 1
Events: 28 Simulation
To simulate the following for the above subject with specific observation time points.
- Plasma concentration.
- Amount excreted unchanged in urine.
Random.seed!(123)The random effects are zero’ed out since we are simulating means
zfx = zero_randeffs(pk_16, sub1, param)(η = [0.0, 0.0, 0.0, 0.0, 0.0],)sim_sub1 = simobs(
pk_16,
sub1,
param,
zfx,
obstimes = [
0.5,
1,
2,
4,
6.1,
7.6,
8.02,
12.05,
12.15,
15.95,
22.13,
23.89,
24.05,
24.46,
24.94,
25.94,
26.96,
27.95,
29.97,
31.94,
35.96,
36.2,
48,
48.2,
54,
60,
60.2,
72,
72.2,
],
)SimulatedObservations
Simulated variables: cp_plasma, dv_plasma, cp_urine, dv_urine
Time: [0.5, 1.0, 2.0, 4.0, 6.1, 7.6, 8.02, 12.05, 12.15, 15.95 … 31.94, 35.96, 36.2, 48.0, 48.2, 54.0, 60.0, 60.2, 72.0, 72.2]9 Visualization
Create a DataFrame of the simulated data.
df1 = DataFrame(sim_sub1)
first(df1, 5)| Row | id | time | cp_plasma | dv_plasma | cp_urine | dv_urine | evid | amt | cmt | rate | duration | ss | ii | route | tad | dosenum | Central | Peripheral | Urine | CLr | CLm | Vc | Vp | Q | CL | Vss | t12 | η_1 | η_2 | η_3 | η_4 | η_5 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| String | Float64 | Float64? | Float64? | Float64? | Float64? | Int64 | Float64? | Symbol? | Float64? | Float64? | Int8? | Float64? | NCA.Route? | Float64 | Int64 | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64 | Float64 | Float64 | Float64 | Float64 | |
| 1 | 1 | 0.0 | missing | missing | missing | missing | 1 | 538.0 | Central | 547.304 | 0.983 | 0 | 0.0 | NullRoute | 0.0 | 1 | missing | missing | missing | 0.31 | 0.05 | 1.6 | 0.16 | 0.03 | 0.36 | 1.76 | 3.388 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 2 | 1 | 0.5 | 161.044 | 150.645 | 12.7335 | 10.8702 | 0 | 0.0 | missing | 0.0 | 0.0 | 0 | 0.0 | missing | 0.5 | 1 | 257.67 | 1.19427 | 12.7335 | 0.31 | 0.05 | 1.6 | 0.16 | 0.03 | 0.36 | 1.76 | 3.388 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 3 | 1 | 0.983 | missing | missing | missing | missing | 1 | 3390.0 | Central | 147.603 | 22.967 | 0 | 0.0 | NullRoute | 0.0 | 2 | missing | missing | missing | 0.31 | 0.05 | 1.6 | 0.16 | 0.03 | 0.36 | 1.76 | 3.388 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 4 | 1 | 1.0 | 299.498 | 250.871 | 48.9648 | 47.899 | 0 | 0.0 | missing | 0.0 | 0.0 | 0 | 0.0 | missing | 0.017 | 2 | 479.197 | 4.45023 | 48.9648 | 0.31 | 0.05 | 1.6 | 0.16 | 0.03 | 0.36 | 1.76 | 3.388 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| 5 | 1 | 2.0 | 317.468 | 333.095 | 144.684 | 158.878 | 0 | 0.0 | missing | 0.0 | 0.0 | 0 | 0.0 | missing | 1.017 | 2 | 507.948 | 12.1436 | 144.684 | 0.31 | 0.05 | 1.6 | 0.16 | 0.03 | 0.36 | 1.76 | 3.388 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
time_plasma = [
0.5,
1,
2,
4,
7.6,
8.02,
12.05,
15.95,
22.13,
23.89,
24.46,
24.94,
25.94,
26.96,
27.95,
29.97,
31.94,
35.96,
48,
54,
60,
72,
]
df_plasma = @rsubset df1 :time in time_plasmatime_urine = [6.1, 12.15, 24.05, 36.2, 48.2, 60.2, 72.2]
df_urine = @rsubset df1 :time in time_urinePlot the simulated plasma concentration data and the amount of drug excreted in urine in a single plot.
plasma_plt = @chain df_plasma begin
@rtransform :Matrix = "Plasma"
data(_) *
mapping(
:time => "Time (hours)",
:cp_plasma => "Concentration (μM) & Amount (μmol)",
color = :Matrix,
) *
visual(ScatterLines, color = :blue, linewidth = 4)
end
urine_plt = @chain df_urine begin
@rtransform :Matrix = "Urine"
data(_) *
mapping(
:time => "Time (hours)",
:cp_urine => "Concentration (μM) & Amount (μmol)",
color = :Matrix,
) *
visual(ScatterLines, color = :black, linewidth = 4)
end
draw(
plasma_plt + urine_plt;
axis = (;
xticks = [0, 10, 20, 30, 40, 50, 60, 70, 80],
yticks = [0.1, 0, 1, 10, 100, 1000, 10000],
yscale = log,
ytickformat = x -> string.(round.(x; digits = 1)),
ygridwidth = 3,
yminorticksvisible = true,
yminorgridvisible = true,
yminorticks = IntervalsBetween(10),
xminorticksvisible = true,
xminorgridvisible = true,
xminorticks = IntervalsBetween(5),
),
figure = (; fontsize = 22,),
)10 Population simulation
par = (
tvclm = 0.05,
tvclr = 0.31,
tvvc = 1.6,
tvvp = 0.16,
tvq = 0.03,
Ω = Diagonal([0.0081, 0.004, 0.0004, 0.0256, 0.0676]),
σ₁_prop = 0.04,
σ₂_prop = 0.09,
)
ev1 = DosageRegimen([538, 3390], time = [0, 0.983], cmt = [1, 1], rate = [547.304, 147.603])
pop = map(i -> Subject(id = i, events = ev1), 1:80)
Random.seed!(1234)
pop_sim = simobs(
pk_16,
pop,
par,
obstimes = [
0.5,
1,
2,
4,
6.1,
7.6,
8.02,
12.05,
15.95,
22.13,
23.89,
24.46,
24.94,
25.94,
26.96,
27.95,
29.97,
31.94,
36,
48,
54,
60,
72,
],
)
df_sim = DataFrame(pop_sim)
#CSV.write("pk_16.csv", df_sim)