using Pumas
using PumasUtilities
using Random
using CairoMakie
using AlgebraOfGraphics
using DataFramesMeta
using CSV
PK51 - Multi-Compartment Drug/Metabolite
1 Learning Outcome
In this tutorial, you will gain a greater appreciation and learn about two compartment parent - metabolite kinetics.
With this modeling exercise, a drug is administered both IV and orally at different occasions to different subjects, and plasma data is collected. The concentrations are obtained for both the drugs and metabolites.
2 Objectives
In this tutorial, you will learn how to build a multi compartment drug/metabolite model and to simulate the model for different subjects and dosage regimens.
3 Background
Before constructing a model, it is important to establish the process the model will follow and a scenario for the simulation.
Below is the scenario for this tutorial:
- Structural model - Multi-Compartment drug/metabolite
- Route of administration - IV administration to one subject and oral administration to another subject
- Dosage Regimen - 5000 mg IV and 8000 mg Oral
- Number of Subjects - 2
This diagram describes how such an administered dose will be handled, which facilitates building the model. 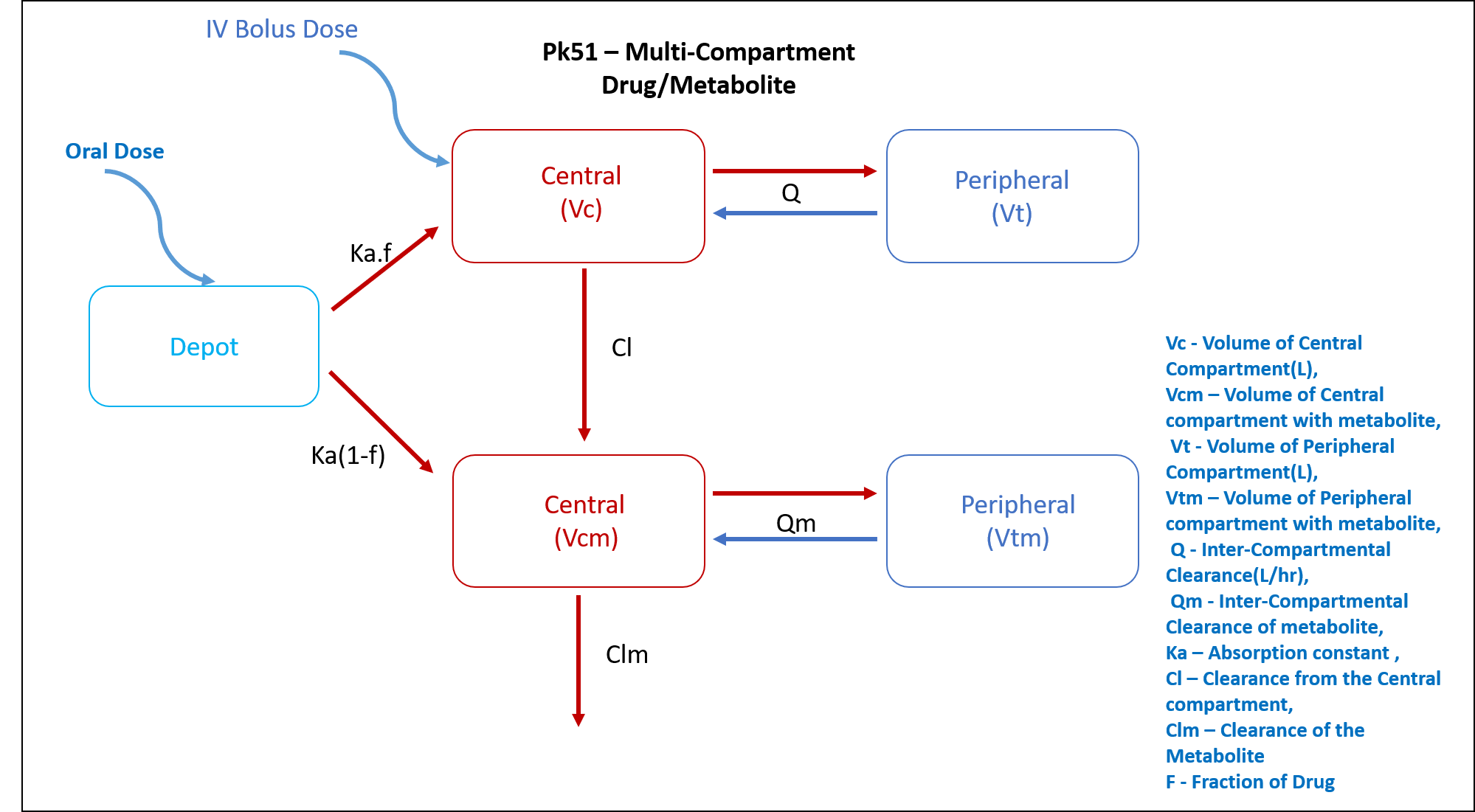
4 Libraries
Call the required libraries to get started.
5 Model
In this two compartment model, we administer the dose to the oral and central compartments on two different occasions.
pk_51 = @model begin
@metadata begin
desc = "Two Compartment Model with Metabolite Compartment"
timeu = u"hr"
end
@param begin
"""
Volume of Central Compartment (L)
"""
tvvc ∈ RealDomain(lower = 0)
"""
Clearance (L/hr)
"""
tvcl ∈ RealDomain(lower = 0)
"""
Inter Compartmental Clearance (L/hr)
"""
tvcld ∈ RealDomain(lower = 0)
"""
Volume of Peripheral Compartment (L)
"""
tvvt ∈ RealDomain(lower = 0)
"""
Volume of Central Compartment of Metabolite (L)
"""
tvvcm ∈ RealDomain(lower = 0)
"""
Clearance of metabolite (L/hr)
"""
tvclm ∈ RealDomain(lower = 0)
"""
Inter Compartmental Clearance of Metabolite (L/hr)
"""
tvcldm ∈ RealDomain(lower = 0)
"""
Volume of Peripheral Compartment of Metabolite (L)
"""
tvvtm ∈ RealDomain(lower = 0)
"""
Absorption rate constant (hr⁻¹)
"""
tvka ∈ RealDomain(lower = 0)
"""
Fraction of drug absorbed
"""
tvf ∈ RealDomain(lower = 0)
"""
Lag time (hr)
"""
tvlag ∈ RealDomain(lower = 0)
Ω ∈ PDiagDomain(7)
"""
Proportional RUV - Plasma
"""
σ²_prop_cp ∈ RealDomain(lower = 0)
"""
Proportional RUV - Metabolite
"""
σ²_prop_met ∈ RealDomain(lower = 0)
end
@random begin
η ~ MvNormal(Ω)
end
@pre begin
Vc = tvvc * exp(η[1])
Cl = tvcl * exp(η[2])
Cld = tvcld * exp(η[3])
Vt = tvvt * exp(η[4])
Vcm = tvvcm
Clm = tvclm * exp(η[5])
Cldm = tvcldm
Vtm = tvvtm * exp(η[6])
Ka = tvka * exp(η[7])
end
@dosecontrol begin
bioav = (Depot = tvf, Metabolite = (1 - tvf))
lags = (Depot = tvlag,)
end
@dynamics begin
Depot' = -Ka * Depot
Central' =
Ka * Depot - (Cl / Vc) * Central - (Cld / Vc) * Central +
(Cld / Vt) * Peripheral
Peripheral' = (Cld / Vc) * Central - (Cld / Vt) * Peripheral
Metabolite' =
Ka * Depot + (Cl / Vc) * Central - (Clm / Vcm) * Metabolite -
(Cldm / Vcm) * Metabolite + (Cldm / Vtm) * PeriMetabolite
PeriMetabolite' = (Cldm / Vcm) * Metabolite - (Cldm / Vtm) * PeriMetabolite
end
@derived begin
cp = @. Central / Vc
met = @. Metabolite / Vcm
"""
Observed Concentration (...)
"""
dv_cp ~ @. Normal(cp, sqrt(cp^2 * σ²_prop_cp))
"""
Observed Concentration (...)
"""
dv_met ~ @. Normal(met, sqrt(met^2 * σ²_prop_met))
end
endPumasModel
Parameters: tvvc, tvcl, tvcld, tvvt, tvvcm, tvclm, tvcldm, tvvtm, tvka, tvf, tvlag, Ω, σ²_prop_cp, σ²_prop_met
Random effects: η
Covariates:
Dynamical system variables: Depot, Central, Peripheral, Metabolite, PeriMetabolite
Dynamical system type: Matrix exponential
Derived: cp, met, dv_cp, dv_met
Observed: cp, met, dv_cp, dv_met6 Parameters
The parameters are as given below.
Cl- Clearance (L/hr)Clm- Clearance of metabolite (L/hr)Cld- Inter Compartmental Clearance (L/hr)Cldm- Inter Compartmental Clearance of metabolite (L/hr)Vc- Volume of Central Compartment (L)Vcm- Volume of Central Compartment of Metabolite (L)f- Fraction of drug absorbedlags- Lag time (hr)Ka- Absorption rate constant (hr⁻¹)Vt- Volume of Peripheral Compartment (L)Vtm- Volume of Peripheral Compartment of metabolite (L)Ω- Between Subject Variabilityσ- Residual error
These are the initial estimates we will be using in this model exercise. Note that tv represents the typical value for parameters.
param = (;
tvvc = 18.7,
tvcl = 0.55,
tvcld = 0.073,
tvvt = 10,
tvvcm = 4.9,
tvclm = 0.08,
tvcldm = 0.58,
tvvtm = 55,
tvka = 0.03,
tvf = 0.24,
tvlag = 21,
Ω = Diagonal([0.01, 0.01, 0.01, 0.01, 0.01, 0.01, 0.01]),
σ²_prop_cp = 0.015,
σ²_prop_met = 0.015,
)7 Dosage Regimen
To start the simulation process, the dosing regimen from the background section must be developed first prior to running a simulation.
7.1 IV
A single dose of 5000 mg given as a rapid IV injection.
The dosage regimen is specified as:
ev1 = DosageRegimen(5000; time = 0, cmt = 2)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 2 | 5000.0 | 1 | 0.0 | 0 | 0.0 | 0.0 | 0 | NullRoute |
This is how to create the single subject undergoing the dosing regimen above.
sub1 = Subject(; id = 1, events = ev1)Subject
ID: 1
Events: 17.2 Oral
A single dose of 8000 mg given orally.
The dosage regimen is specified as:
ev2 = DosageRegimen(8000; time = 0, cmt = 1)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 1 | 8000.0 | 1 | 0.0 | 0 | 0.0 | 0.0 | 0 | NullRoute |
This is how to create the single subject undergoing the dosing regimen above.
sub2 = Subject(; id = 2, events = ev2)Subject
ID: 2
Events: 1This is how to put both subjects together into a population.
pop2_sub = [sub1, sub2]Population
Subjects: 2
Observations: 8 Simulation
Simulate using the simobs function.
Random.seed!()
The Random.seed! function is included here for purposes of reproducibility of the simulation in this tutorial. Specification of a seed value would not be required in a Pumas workflow that is estimating model parameters.
Random.seed!(123)sim_pop2_sub = simobs(pk_51, pop2_sub, param, obstimes = 0.1:0.1:1500)Simulated population (Vector{<:Subject})
Simulated subjects: 2
Simulated variables: cp, met, dv_cp, dv_met9 Visualization
In the following plots below, we can see the concentration profiles of the parent drug and the metabolite.
@chain DataFrame(sim_pop2_sub) begin
dropmissing(:cp)
data(_) *
mapping(
:time => "Time (hours)",
:cp => "Parent Concentration (-)";
color = :id => nonnumeric => "ID",
) *
visual(Lines; linewidth = 4)
draw(; figure = (; fontsize = 22), axis = (; xticks = 0:150:1500))
end@chain DataFrame(sim_pop2_sub) begin
dropmissing(:met)
data(_) *
mapping(
:time => "Time (hours)",
:met => "Metabolite Concentration (-)";
color = :id => nonnumeric => "ID",
) *
visual(Lines; linewidth = 4)
draw(; figure = (; fontsize = 22), axis = (; xticks = 0:150:1500))
end10 Population Simulation
This block updates the parameters of the model to increase intersubject variability in parameters and defines timepoints for prediction of concentrations. The results are written to a CSV file.
Creating a larger population for the simulation and utilizing the following initial estimates for the simulation below:
par = (
tvvc = 18.7,
tvcl = 0.55,
tvcld = 0.073,
tvvt = 10,
tvvcm = 4.9,
tvclm = 0.08,
tvcldm = 0.58,
tvvtm = 55,
tvka = 0.03,
tvf = 0.24,
tvlag = 21,
Ω = Diagonal([0.04, 0.02, 0.02, 0.03, 0.02, 0.04, 0.04]),
σ²_prop_cp = 0.04,
σ²_prop_met = 0.09,
)
ev1 = DosageRegimen(5000, time = 0, cmt = 2)
pop1 = map(i -> Subject(id = i, events = ev1), 1:25)
ev2 = DosageRegimen(8000, time = 0, cmt = 1)
pop2 = map(i -> Subject(id = 1, events = ev2), 26:50)Simulating the population and obtaining the results.
pop = [pop1; pop2]
Random.seed!(1234)
sim_pop = simobs(
pk_51,
pop,
par,
obstimes = [2, 5, 10, 15, 30, 45, 60, 90, 120, 180, 240, 360, 480, 720, 1440],
)
sim_plot(sim_pop)
df_sim = DataFrame(sim_pop)
#CSV.write("pk51.csv", df_sim);With the CSV.write function, you can input the name of the dataframe (df_sim) and the file name of your choice (pk_51.csv) to save the file to your local directory or repository.
11 Conclusion
Constructing a multi-compartment model involves:
- understanding the process of how the drug and metabolite are passed through the two-compartment system,
- translating processes into ODEs using Pumas, and
- simulating the model in a single patient for evaluation.