using Random
using Pumas
using PumasUtilities
using AlgebraOfGraphics
using CairoMakie
using CSV
using DataFramesMeta
using Dates
PK24 - Non-linear Kinetics - Flow II
1 Background
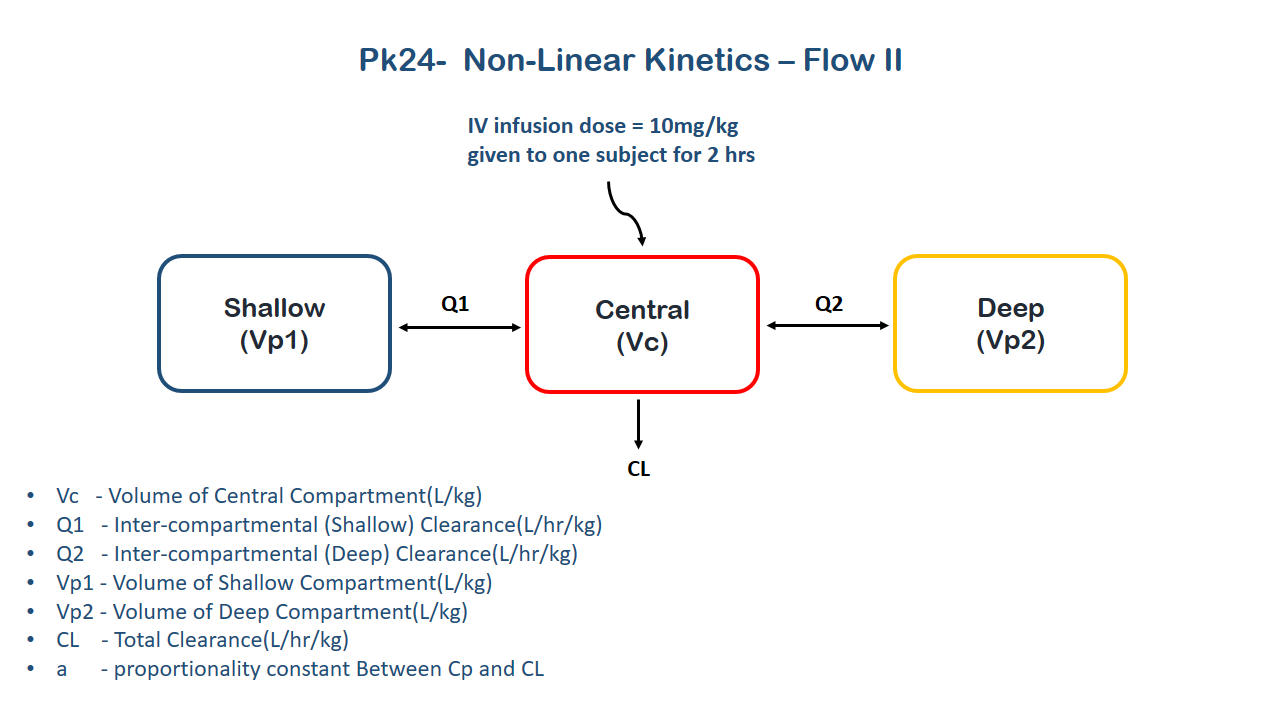
- Structural model - Multi-compartment (three) model with concentration dependent clearance
- Route of administration - IV infusion
- Dosage Regimen - One IV infusion dose of 10 mg/kg given for 2 hours.
- Number of Subjects - 1
2 Learning Outcomes
This exercise explains a multi-compartment model following concentration dependent clearance.
3 Objectives
In this tutorial, you will learn:
- To build a multi-compartment model with concentration dependent clearance.
- To simulate the data for one subject given an IV infusion for 2 hours.
4 Libraries
Call the necessary libraries to get started.
5 Model
This model contains three compartments (Central, shallow and deep) and the clearance is dependent on the change in plasma concentration due to the drug, which is known to reduce cardiac output and hepatic blood flow with an increase in plasma concentration. The change in clearance can be accounted by the equation: \(CL= CLo-A(Central/Vc)\) where \(A\) is proportionality constant between \(CL\) and \(C\)
pk_24 = @model begin
@metadata begin
desc = "Three Compartment Model"
timeu = u"hr"
end
@param begin
"""
Volume of Central Compartment(L/kg)
"""
tvvc ∈ RealDomain(lower = 0)
"""
Clearance(L/hr/kg)
"""
tvclo ∈ RealDomain(lower = 0)
"""
Inter-compartmental (Shallow) Clearance(L/hr/kg)
"""
tvq1 ∈ RealDomain(lower = 0)
"""
Inter-compartmental (Deep) Clearance(L/hr/kg)
"""
tvq2 ∈ RealDomain(lower = 0)
"""
Volume of Shallow Compartment(L/kg)
"""
tvvp1 ∈ RealDomain(lower = 0)
"""
Volume of Deep Compartment(L/kg)
"""
tvvp2 ∈ RealDomain(lower = 0)
"""
Proportionality constant Between Cp and CL(L2/hr/μg/kg)
"""
tva ∈ RealDomain(lower = 0)
Ω ∈ PDiagDomain(7)
σ_prop ∈ RealDomain(lower = 0)
end
@random begin
η ~ MvNormal(Ω)
end
@pre begin
Vc = tvvc * exp(η[1])
CLo = tvclo * exp(η[2])
Q1 = tvq1 * exp(η[3])
Q2 = tvq2 * exp(η[4])
Vp1 = tvvp1 * exp(η[5])
Vp2 = tvvp2 * exp(η[6])
A = 1000 * tva * exp(η[7])
end
@dynamics begin
Central' =
-(CLo - (A * (Central / Vc))) * (Central / Vc) - Q1 * (Central / Vc) +
Q1 * (Shallow / Vp1) - Q2 * (Central / Vc) + Q2 * (Deep / Vp2)
Shallow' = Q1 * (Central / Vc) - Q1 * (Shallow / Vp1)
Deep' = Q2 * (Central / Vc) - Q2 * (Deep / Vp2)
end
@derived begin
cp = @. 1000 * Central / Vc
"""
Observed Concentration (μg/L)
"""
dv ~ @. ProportionalNormal(cp, σ_prop)
end
end┌ Warning: Variable `cp` is defined in the `@derived` block using `=` and hence `cp` is not used for model fitting but only returned when simulating: │ If `cp` is a random variable, it must be defined in the `@derived` block using `~`; │ if `cp` should be returned when simulating, it should be defined in the `@observed` block using `=`; │ if `cp` is an intermediate quantity that should not be returned when simulating, it should be defined using `:=`. └ @ Pumas ~/run/_work/PumasTutorials.jl/PumasTutorials.jl/custom_julia_depot/packages/Pumas/qxx5c/src/dsl/model_macro.jl:2351
PumasModel
Parameters: tvvc, tvclo, tvq1, tvq2, tvvp1, tvvp2, tva, Ω, σ_prop
Random effects: η
Covariates:
Dynamical system variables: Central, Shallow, Deep
Dynamical system type: Nonlinear ODE
Derived: cp, dv
Observed: cp, dv6 Parameters
The parameters are as given below. Note that tv represents the typical value for parameters.
Vc- Volume of Central Compartment (L/kg)CLo- Clearance (L/hr/kg)Q1- Inter-compartmental (Shallow) Clearance (L/hr/kg)Q2- Inter-compartmental (Deep) Clearance (L/hr/kg)Vp1- Volume of Shallow Compartment (L/kg)Vp2- Volume of Deep Compartment (L/kg)A- proportionality constant Between Cp and CL (L2/hr/μg/kg)Ω- Between Subject Variabilityσ- Residual error (for plasma concentration)
param = (
tvvc = 0.68,
tvclo = 6.61,
tvq1 = 5.94,
tvq2 = 0.93,
tvvp1 = 1.77,
tvvp2 = 3.20,
tva = 0.0025,
Ω = Diagonal([0.04, 0.04, 0.04, 0.04, 0.04, 0.04, 0.04]),
σ_prop = 0.004,
)7 Dosage Regimen
Build the dosage regimen for one subject given an IV infusion of 10 mg/kg for 2 hours.
DR = DosageRegimen(10, time = 0, cmt = 1, duration = 2)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 1 | 10.0 | 1 | 0.0 | 0 | 5.0 | 2.0 | 0 | NullRoute |
s1 = Subject(id = 1, events = DR)Subject
ID: 1
Events: 18 Simulation
The random effects are zero’ed out since we are simulating means
zfx = zero_randeffs(pk_24, s1, param)(η = [0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0],)To simulate the data with specific data points.
sim_sub1 = simobs(pk_24, s1, param, zfx, obstimes = 0.1:0.01:8)SimulatedObservations
Simulated variables: cp, dv
Time: 0.1:0.01:8.09 Visualization
@chain DataFrame(sim_sub1) begin
dropmissing(:cp)
data(_) *
mapping(:time => "Time (hour)", :cp => "PK24 Concentrations (μg/mL)") *
visual(Lines, linewidth = 4)
draw(; axis = (; xticks = 0:2:8), figure = (; fontsize = 22))
end10 Population simulation
parameters = (
tvvc = 0.68,
tvclo = 6.61,
tvq1 = 5.94,
tvq2 = 0.93,
tvvp1 = 1.77,
tvvp2 = 3.20,
tva = 0.0025,
Ω = Diagonal([0.02, 0.0, 0.02, 0.04, 0.02, 0.02, 0.0]),
σ_prop = 0.039,
)
DR = DosageRegimen(10, time = 0, cmt = 1, duration = 2)
pop = map(i -> Subject(id = i, events = DR), 1:45)
Random.seed!(1234)
ss = simobs(
pk_24,
pop,
parameters,
obstimes = [
0.25,
0.5,
0.75,
1.05,
1.25,
1.49,
1.75,
1.99,
2.16,
2.35,
2.4,
2.65,
2.81,
2.95,
3.11,
3.56,
4.15,
6,
7,
],
)
df_sim = DataFrame(ss)
#CSV.write("pk_24.csv", df_sim)