using Pumas
using PumasUtilities
using AlgebraOfGraphics
using CairoMakie
using ColorSchemes
using Random
using CSV
using DataFramesMeta
using Dates
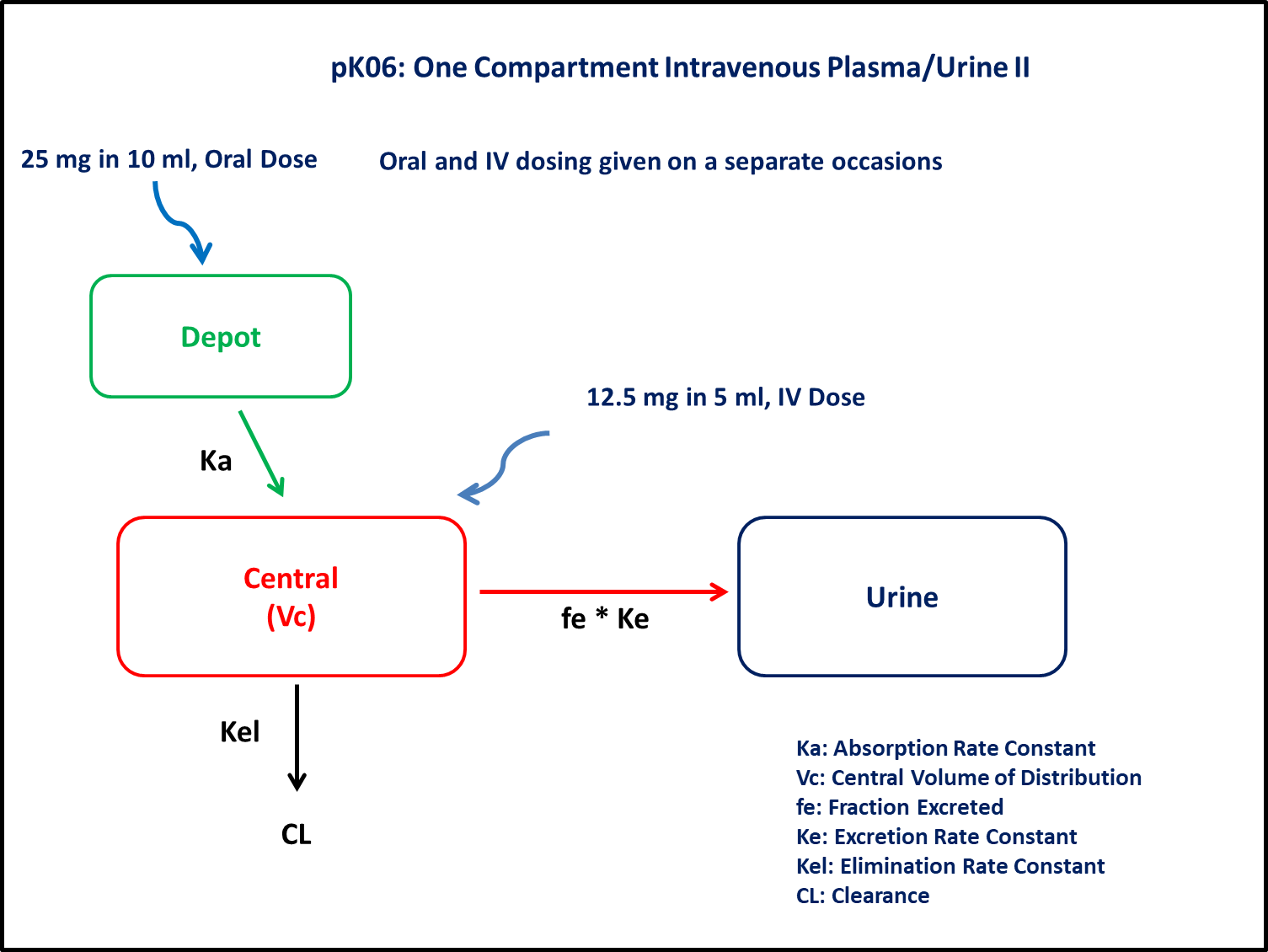
PK06 - One-compartment intravenous plasma/urine II
1 Background
- Structural model - One compartment linear elimination with first order absorption
- Route of administration - Oral and IV given simultaneously
- Dosage Regimen - 12.5 mg IV and 25 mg Oral
- Number of Subjects - 1
2 Objective
In this tutorial, you will learn to build a one compartment model and to simulate the model for different subjects and dosage regimens.
3 Libraries
Call the necessary libraries to get started
4 Model
In this one compartment model, we administer doses both oral and IV on two different occasions.
pk_06 = @model begin
@metadata begin
desc = "One Compartment Model with Urine Cmt"
timeu = u"hr"
end
@param begin
"""
Clearance (L/hr)
"""
tvcl ∈ RealDomain(lower = 0)
"""
Volume of Distribution (L)
"""
tvvc ∈ RealDomain(lower = 0)
"""
Fraction of drug excreted in Urine
"""
tvfe ∈ RealDomain(lower = 0)
"""
Absorption Rate Constant (1/hr)
"""
tvka ∈ RealDomain(lower = 0)
"""
Lag-time (hr)
"""
tvlag ∈ RealDomain(lower = 0)
"""
Bioavailability
"""
tvF ∈ RealDomain(lower = 0)
Ω ∈ PDiagDomain(6)
"""
Proportional RUV
"""
σ²_prop ∈ RealDomain(lower = 0)
"""
Additive RUV
"""
σ_add ∈ RealDomain(lower = 0)
end
@random begin
η ~ MvNormal(Ω)
end
@pre begin
Cl = tvcl * exp(η[1])
Vc = tvvc * exp(η[2])
Ka = tvka * exp(η[3])
fe = tvfe * exp(η[4])
end
@dosecontrol begin
lags = (Depot = tvlag * exp(η[5]),)
bioav = (Depot = tvF * exp(η[6]),)
end
@dynamics begin
Depot' = -Ka * Depot
Central' = Ka * Depot - (Cl / Vc) * Central
Urine' = fe * (Cl / Vc) * Central
end
@derived begin
"""
PK06 Plasma Concentration (μg/L)
"""
cp = @. (Central / Vc)
dv ~ @. Normal(cp, sqrt(cp^2 * σ²_prop))
"""
PK06 Urine Amount (μg)
"""
ae = @. Urine
dv_ae ~ @. Normal(ae, σ_add)
end
endPumasModel
Parameters: tvcl, tvvc, tvfe, tvka, tvlag, tvF, Ω, σ²_prop, σ_add
Random effects: η
Covariates:
Dynamical system variables: Depot, Central, Urine
Dynamical system type: Matrix exponential
Derived: cp, dv, ae, dv_ae
Observed: cp, dv, ae, dv_ae5 Parameters
Parameters provided for simulation. Note that tv represents the typical value for parameters.
Cl- Clearance (L/hr)Vc- Volume of Central Compartment (L)Fe- Fraction of drug excreted in Urinelag- lag time (hr)Ka- Absorption rate constant (hr⁻¹)F- BioavailabilityΩ- Between Subject Variabilityσ- Residual error
param = (
tvcl = 6.02527,
tvvc = 290.292,
tvfe = 0.0698294,
tvka = 0.420432,
tvlag = 0.311831,
tvF = 1.13591,
Ω = Diagonal([0.04, 0.04, 0.04, 0.04, 0.04, 0.04]),
σ²_prop = 0.015,
σ_add = 3,
)6 Dosage Regimen
6.1 For IV
A single subject receives an IV-bolus dose of 12.5 mg or 12500 μg
ev1 = DosageRegimen(12500, time = 0, cmt = 2)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 2 | 12500.0 | 1 | 0.0 | 0 | 0.0 | 0.0 | 0 | NullRoute |
sub1 = Subject(id = "1 - IV", events = ev1)Subject
ID: 1 - IV
Events: 16.2 For Oral
A single subject receives an oral dose of 25 mg or 25000 μg
ev2 = DosageRegimen(25000, time = 0, cmt = 1)| Row | time | cmt | amt | evid | ii | addl | rate | duration | ss | route |
|---|---|---|---|---|---|---|---|---|---|---|
| Float64 | Int64 | Float64 | Int8 | Float64 | Int64 | Float64 | Float64 | Int8 | NCA.Route | |
| 1 | 0.0 | 1 | 25000.0 | 1 | 0.0 | 0 | 0.0 | 0.0 | 0 | NullRoute |
sub2 = Subject(id = "2 - PO", events = ev2)Subject
ID: 2 - PO
Events: 1pop = [sub1, sub2]Population
Subjects: 2
Observations: 7 Simulation
Here, we will learn how to simultaneously simulate plasma concentration and urine amount after IV and oral administration.
Random.seed!(123)The random effects are zero’ed out since we are simulating means
zfx = zero_randeffs(pk_06, pop, param)2-element Vector{NamedTuple{(:η,), Tuple{Vector{Float64}}}}:
(η = [0.0, 0.0, 0.0, 0.0, 0.0, 0.0],)
(η = [0.0, 0.0, 0.0, 0.0, 0.0, 0.0],)sim = simobs(pk_06, pop, param, zfx, obstimes = 0.1:0.1:168)Simulated population (Vector{<:Subject})
Simulated subjects: 2
Simulated variables: cp, dv, ae, dv_ae7.1 For IV
Let’s simulate for plasma concentration and amount excreted unchanged in urine.
The random effects are zero’ed out since we are simulating means
zfx = zero_randeffs(pk_06, sub1, param)(η = [0.0, 0.0, 0.0, 0.0, 0.0, 0.0],)sim_sub1_iv = simobs(pk_06, sub1, param, zfx, obstimes = 0.1:0.1:168)SimulatedObservations
Simulated variables: cp, dv, ae, dv_ae
Time: 0.1:0.1:168.0df_sim_iv = DataFrame(sim_sub1_iv)
first(df_sim_iv, 5)| Row | id | time | cp | dv | ae | dv_ae | evid | lags_Depot | bioav_Depot | amt | cmt | rate | duration | ss | ii | route | η_1 | η_2 | η_3 | η_4 | η_5 | η_6 | Depot | Central | Urine | Cl | Vc | Ka | fe |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| String? | Float64 | Float64? | Float64? | Float64? | Float64? | Int64? | Float64? | Float64? | Float64? | Symbol? | Float64? | Float64? | Int8? | Float64? | NCA.Route? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | |
| 1 | 1 - IV | 0.0 | missing | missing | missing | missing | 1 | 0.311831 | 1.13591 | 12500.0 | Central | 0.0 | 0.0 | 0 | 0.0 | NullRoute | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | missing | missing | missing | 6.02527 | 290.292 | 0.420432 | 0.0698294 |
| 2 | 1 - IV | 0.1 | 42.9708 | 51.894 | 1.80984 | -0.410209 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12474.1 | 1.80984 | 6.02527 | 290.292 | 0.420432 | 0.0698294 |
| 3 | 1 - IV | 0.2 | 42.8817 | 41.8845 | 3.61592 | 0.041409 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12448.2 | 3.61592 | 6.02527 | 290.292 | 0.420432 | 0.0698294 |
| 4 | 1 - IV | 0.3 | 42.7928 | 49.7791 | 5.41826 | 1.41143 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12422.4 | 5.41826 | 6.02527 | 290.292 | 0.420432 | 0.0698294 |
| 5 | 1 - IV | 0.4 | 42.7041 | 49.2717 | 7.21686 | 7.58476 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 12396.7 | 7.21686 | 6.02527 | 290.292 | 0.420432 | 0.0698294 |
7.2 For Oral
Let’s simulate the plasma concentration and amount excreted unchanged in urine.
The random effects are zero’ed out since we are simulating means
zfx = zero_randeffs(pk_06, sub2, param)(η = [0.0, 0.0, 0.0, 0.0, 0.0, 0.0],)sim_sub2_oral = simobs(pk_06, sub2, param, obstimes = 0.1:0.1:168)SimulatedObservations
Simulated variables: cp, dv, ae, dv_ae
Time: 0.1:0.1:168.0df_sim_oral = DataFrame(sim_sub2_oral)
first(df_sim_oral, 5)| Row | id | time | cp | dv | ae | dv_ae | evid | lags_Depot | bioav_Depot | amt | cmt | rate | duration | ss | ii | route | η_1 | η_2 | η_3 | η_4 | η_5 | η_6 | Depot | Central | Urine | Cl | Vc | Ka | fe |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| String? | Float64 | Float64? | Float64? | Float64? | Float64? | Int64? | Float64? | Float64? | Float64? | Symbol? | Float64? | Float64? | Int8? | Float64? | NCA.Route? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | Float64? | |
| 1 | 2 - PO | 0.0 | missing | missing | missing | missing | 1 | 0.297778 | 1.53409 | 25000.0 | Depot | 0.0 | 0.0 | 0 | 0.0 | NullRoute | 0.381318 | -0.0635945 | -0.268354 | -0.227323 | -0.0461145 | 0.3005 | missing | missing | missing | 8.82228 | 272.406 | 0.321478 | 0.0556305 |
| 2 | 2 - PO | 0.1 | 0.0 | 0.0 | 0.0 | -5.18854 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.381318 | -0.0635945 | -0.268354 | -0.227323 | -0.0461145 | 0.3005 | 0.0 | 0.0 | 0.0 | 8.82228 | 272.406 | 0.321478 | 0.0556305 |
| 3 | 2 - PO | 0.2 | 0.0 | 0.0 | 0.0 | -4.76031 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.381318 | -0.0635945 | -0.268354 | -0.227323 | -0.0461145 | 0.3005 | 0.0 | 0.0 | 0.0 | 8.82228 | 272.406 | 0.321478 | 0.0556305 |
| 4 | 2 - PO | 0.3 | 0.100549 | 0.127716 | 5.48435e-5 | 5.53784 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.381318 | -0.0635945 | -0.268354 | -0.227323 | -0.0461145 | 0.3005 | 38324.7 | 27.3902 | 5.48435e-5 | 8.82228 | 272.406 | 0.321478 | 0.0556305 |
| 5 | 2 - PO | 0.4 | 4.54393 | 4.01653 | 0.114672 | -4.90659 | 0 | missing | missing | missing | missing | missing | missing | missing | missing | missing | 0.381318 | -0.0635945 | -0.268354 | -0.227323 | -0.0461145 | 0.3005 | 37112.3 | 1237.79 | 0.114672 | 8.82228 | 272.406 | 0.321478 | 0.0556305 |
@rsubset! df_sim_oral :time > 0.28 Visualization
Combined Plot of IV data and Oral data
plasma_times = [0.3, 0.6, 1, 2, 3, 4, 6, 8, 24, 48, 72, 96, 168]
df_iv_plasma = @rsubset df_sim_iv :time in plasma_times
df_oral_plasma = @rsubset df_sim_oral :time in plasma_timesurine_times = [24, 48]
df_iv_urine = @rsubset df_sim_iv :time in urine_times
df_oral_urine = @rsubset df_sim_oral :time in urine_timesivsim_plt = @chain df_sim_iv begin
@rtransform :Type = "IV Prediction"
dropmissing(:cp)
data(_) *
mapping(
:time => "Time (hours)",
:cp => "Concentration (μg/L) & Amount (μg)",
color = :Type,
) *
visual(Lines, linewidth = 4)
end
iv_plt = @chain df_iv_plasma begin
@rtransform :Type = "IV Observations"
dropmissing(:cp)
data(_) *
mapping(
:time => "Time (hours)",
:cp => "Concentration (μg/L) & Amount (μg)",
color = :Type,
) *
visual(Scatter, color = :blue, markersize = 14)
end
ivurine_plt = @chain df_iv_urine begin
@rtransform :Type = "IV Observations Amount"
dropmissing(:ae)
data(_) *
mapping(
:time => "Time (hours)",
:ae => "Concentration (μg/L) & Amount (μg)",
color = :Type,
) *
visual(ScatterLines, color = :blue, markersize = 14, linewidth = 4)
end
oralsim_plt = @chain df_sim_oral begin
@rtransform :Type = "Oral Prediction"
dropmissing(:cp)
data(_) *
mapping(
:time => "Time (hours)",
:cp => "Concentration (μg/L) & Amount (μg)",
color = :Type,
) *
visual(Lines, linewidth = 4)
end
oral_plt = @chain df_oral_plasma begin
@rtransform :Type = "Oral Observations"
dropmissing(:cp)
data(_) *
mapping(
:time => "Time (hours)",
:cp => "Concentration (μg/L) & Amount (μg)",
color = :Type,
) *
visual(Scatter, markersize = 14)
end
oralurine_plt = @chain df_oral_urine begin
@rtransform :Type = "Oral Observations Amount"
dropmissing(:ae)
data(_) *
mapping(
:time => "Time (hours)",
:ae => "Concentration (μg/L) & Amount (μg)",
color = :Type,
) *
visual(ScatterLines, markersize = 14, linewidth = 4)
end
draw(
ivsim_plt + iv_plt + ivurine_plt + oralsim_plt + oral_plt + oralurine_plt;
axis = (;
xticks = [0, 20, 40, 60, 80, 100, 120, 140, 160, 180],
yscale = Makie.Symlog10(10),
yticks = [1, 10, 100, 1000, 2000],
ytickformat = x -> string.(round.(x; digits = 1)),
ygridwidth = 3,
yminorticksvisible = true,
yminorgridvisible = true,
yminorticks = IntervalsBetween(10),
xminorticksvisible = true,
xminorgridvisible = true,
xminorticks = IntervalsBetween(5),
),
figure = (; fontsize = 22),
legend = (position = :bottom, nbanks = 3, tellwidth = true),
palettes = (; color = ColorSchemes.tableau_10.colors),
)